Absolute Copy Number Estimation using low-coverage whole genome sequencing data - tgac-vumc/ACE

Copy number analysis by low coverage whole genome sequencing using ultra low-input DNA from formalin-fixed paraffin embedded tumor tissue, Genome Medicine
GitHub - Nealelab/whole_genome_analysis_pipeline

Comprehensive Assessment of Somatic Copy Number Variation Calling Using Next-Generation Sequencing Data

GitHub - Nealelab/whole_genome_analysis_pipeline
DNA copy number profiling: from bulk tissue to single cells

American Society for Clinical Pharmacology and Therapeutics - 2019 - Clinical Pharmacology & Therapeutics - Wiley Online Library
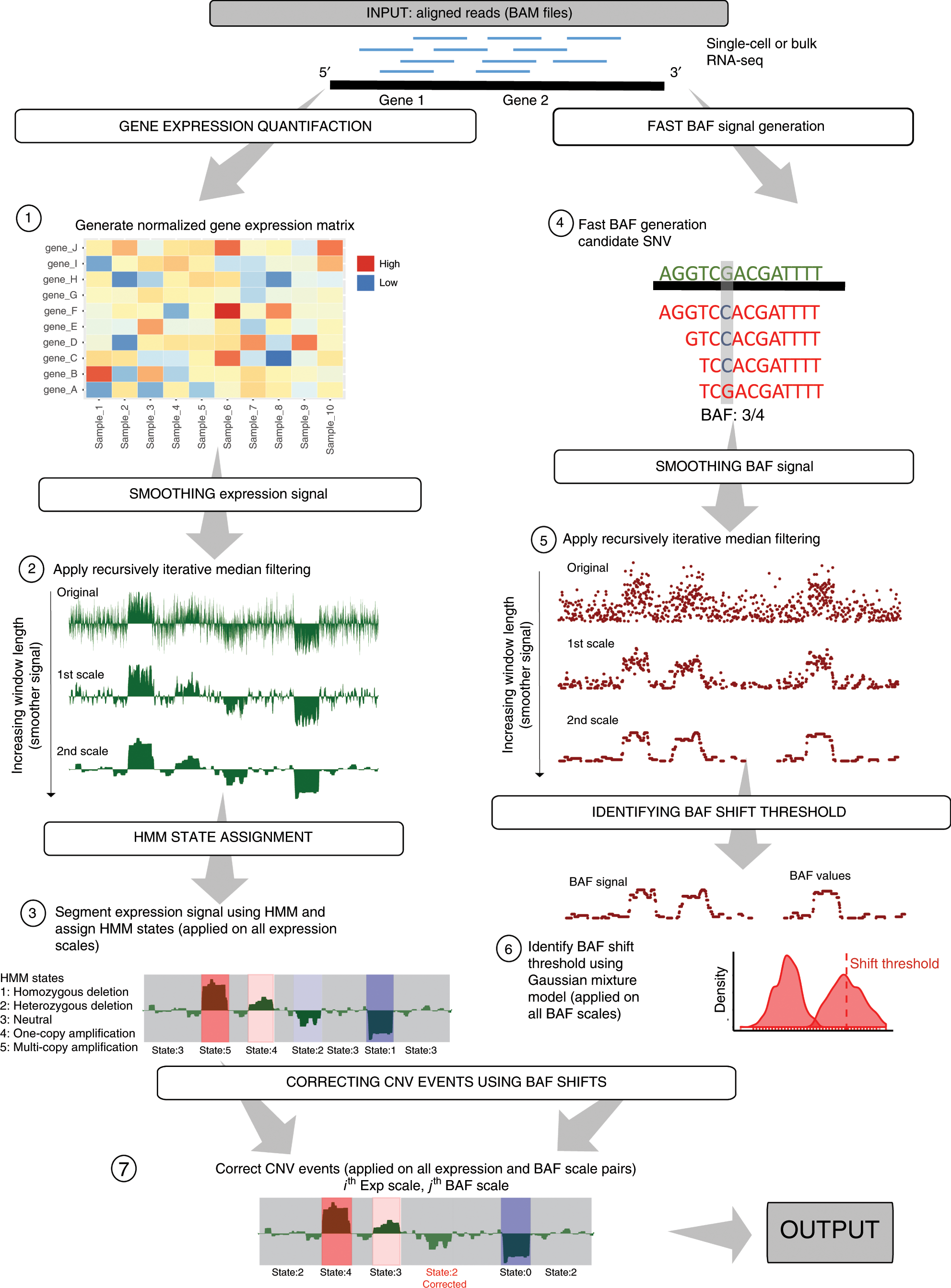
CaSpER identifies and visualizes CNV events by integrative analysis of single-cell or bulk RNA-sequencing data

Shallow whole-genome sequencing of plasma cell-free DNA accurately differentiates small from non-small cell lung carcinoma, Genome Medicine

Assessing Copy Number Alterations in Targeted, Amplicon-Based Next-Generation Sequencing Data - ScienceDirect

Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data
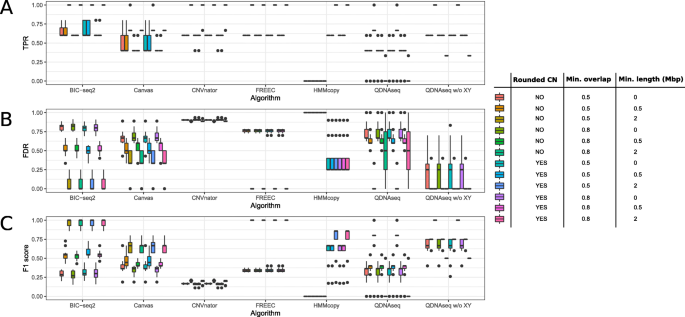
Evaluation of tools for identifying large copy number variations from ultra- low-coverage whole-genome sequencing data, BMC Genomics

PDF] FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing

PDF) Low-Coverage Whole Genome Sequencing Using Laser Capture Microscopy with Combined Digital Droplet PCR: An Effective Tool to Study Copy Number and Kras Mutations in Early Lung Adenocarcinoma Development